2 Input data
In this chapter, we load and preprocess the datasets we use in this study
| Dataset | Reference |
|---|---|
| A dataset of more than 11,000 single cancer and immune cells, classified by cell type | Schelker et al. (2017) |
| 50 Immune cell reference samples from 5 studies | Curated by Finotello et al. (2017) |
| 3 ovarian cancer ascites samples (RNAseq + FACS) | Schelker et al. (2017) |
| 8 PBMC samples (RNAseq + FACS) | Hoek et al. (2015) |
| 4 metastatic melanoma samplese (RNAseq + FACS) | Racle et al. (2017) |
2.1 cell type hierarchy
We use a hierachy of immune cell types to map the cell types between different methods and datasets. The following figure shows this hierarchy visualized as a tree
Figure 2.1: Hierarchy of immune cell types used for mapping cell types between methods and datasets.
2.2 Single cell data for simulated mixtures
In this study, we make use of the single cell dataset curated by Schelker et al. (2017). They aggregated single cell sequencing data from different sources resulting in a set of more than 11,000 single cells. They classified the cells using at set of 45 marker genes into 12 categories:
- 2 cancer types (Melanoma cell, Ovarian carcinoma cell),
- 7 immune cells (B cell, T cell CD8+, T cell CD4+ (non-regulatory), Macrophage/Monocyte, T cell regulatory (Tregs), Dendritic cell, NK cell),
- 2 other cells (Cancer associated fibroblast, Endothelial cell) and
- Unknown cells which could not have been classified unambiguously.
Unknown cells are excluded from the downstream analysis.
The dataset consists of single cells from PBMC, melanoma and ovarian cancer ascites. As we are interested in the deconvolution of cancer samples, we exclude the PBMC cells from all downstream analyses.
| cell_type | n |
|---|---|
| B cell | 646 |
| Cancer associated fibroblast | 132 |
| Dendritic cell | 140 |
| Endothelial cell | 71 |
| Macrophage/Monocyte | 2227 |
| Melanoma cell | 1310 |
| NK cell | 198 |
| Ovarian carcinoma cell | 300 |
| PBMC | 3942 |
| T cell CD4+ (non-regulatory) | 1196 |
| T cell CD8+ | 1130 |
| T cell regulatory (Tregs) | 142 |
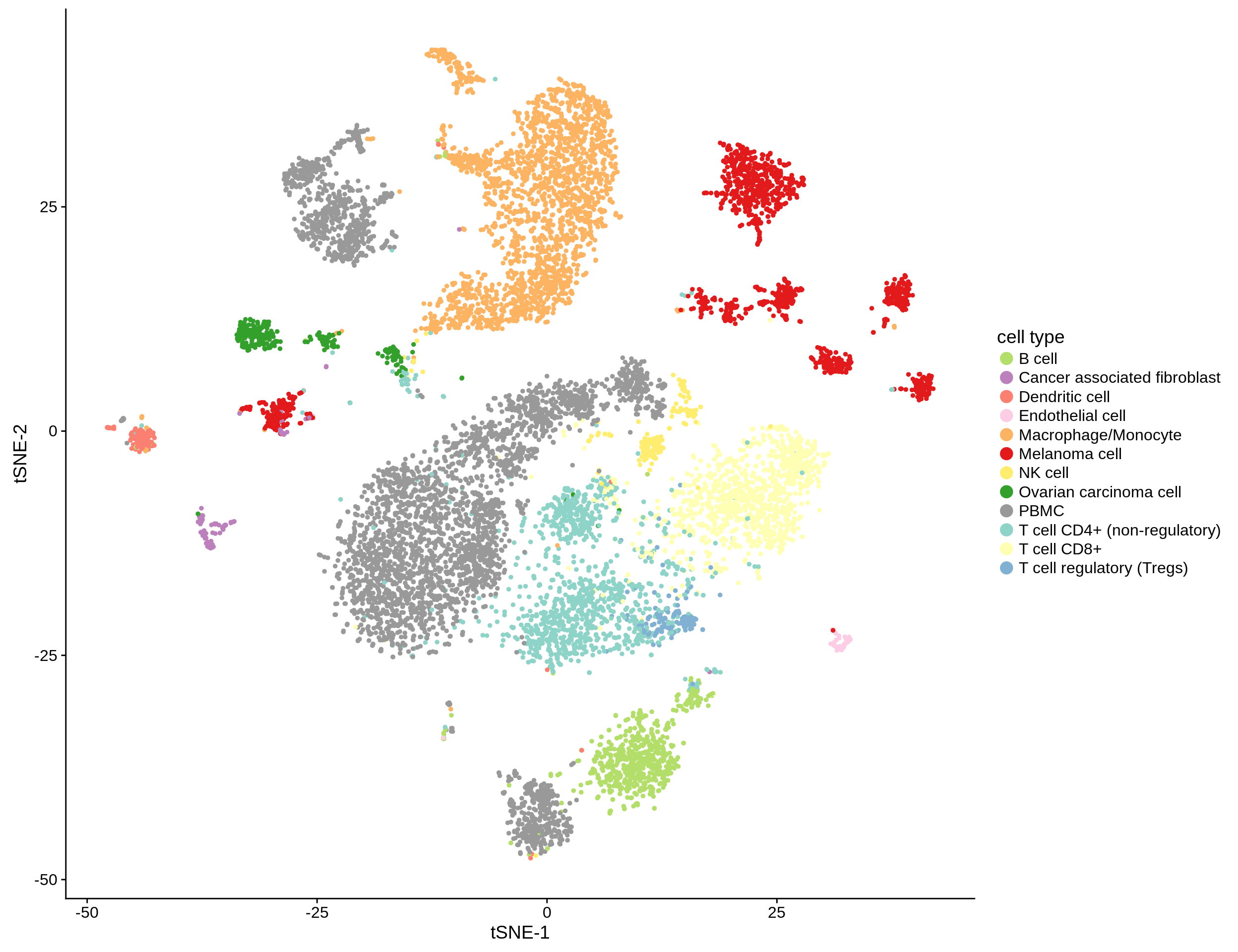
Figure 2.2: tSNE-clustering of the ~12,000 single cells from Schelker et al. (2017).
2.3 Immune cell reference samples
RNAseq samples of pure immune cells of 10 types from 5 studies curated by Finotello et al. (2017).
2.4 8 PBMC samples from Hoek et al. (2015)
2.5 3 ovarian cancer ascites samples from Schelker et al. (2017)
Each sample has two technical replicates. We merge the two replicates by taking the mean for each gene.
The samples have also been profiled by single cell RNA sequencing. The following table shows the cell count for each sample.
| donor | sum(cell_count) |
|---|---|
| 7873M | 864 |
| 7882M | 902 |
| 7892M | 773 |
2.6 4 metastatic melanoma samples from Racle et al. (2017)
2.7 Data sanity checks
Here, we plot the distributions of the different gene expression datasets to ensure that everything looks like we expect it to, e.g. if all datasets are on non-log scale.
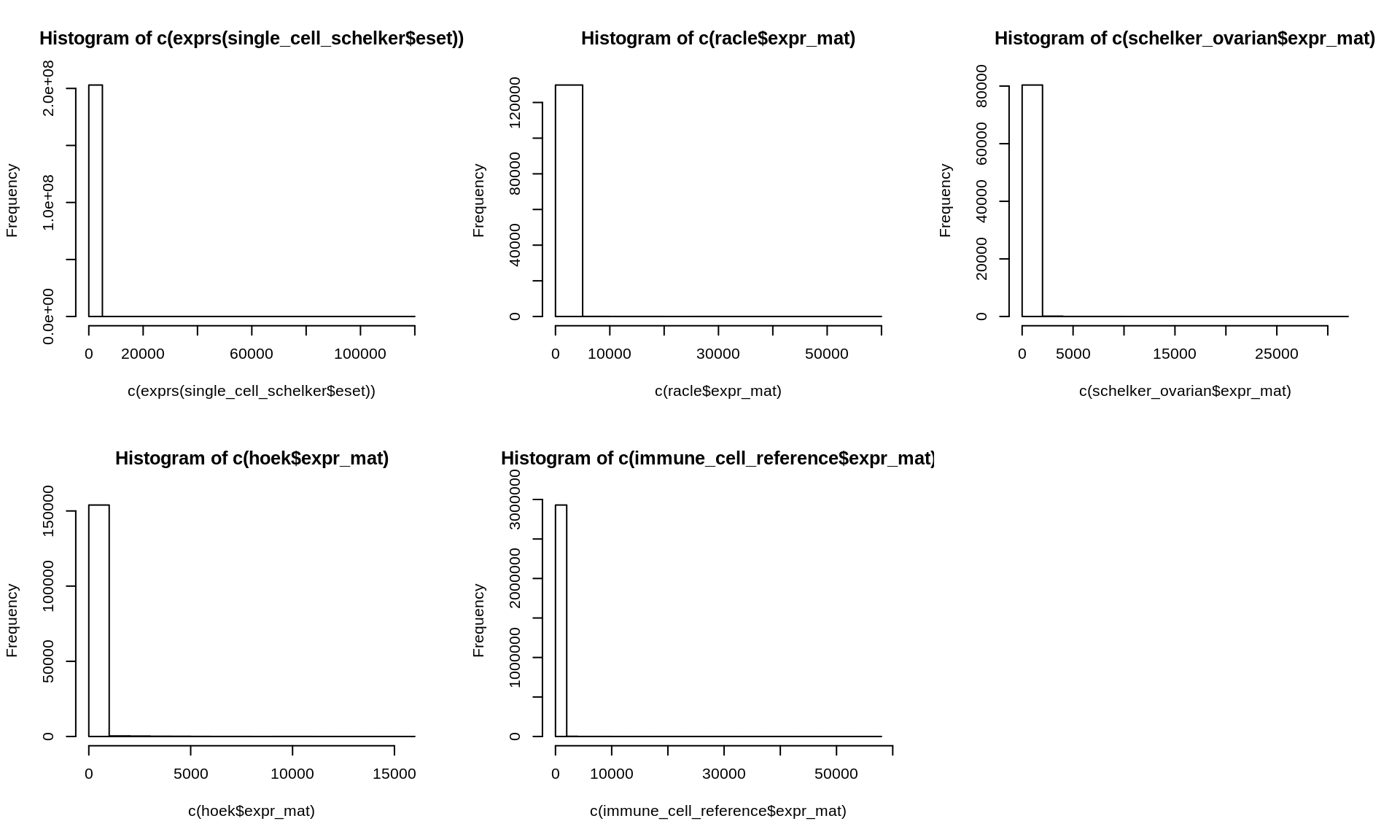
Figure 2.3: Histogram of gene expression data of all datasets
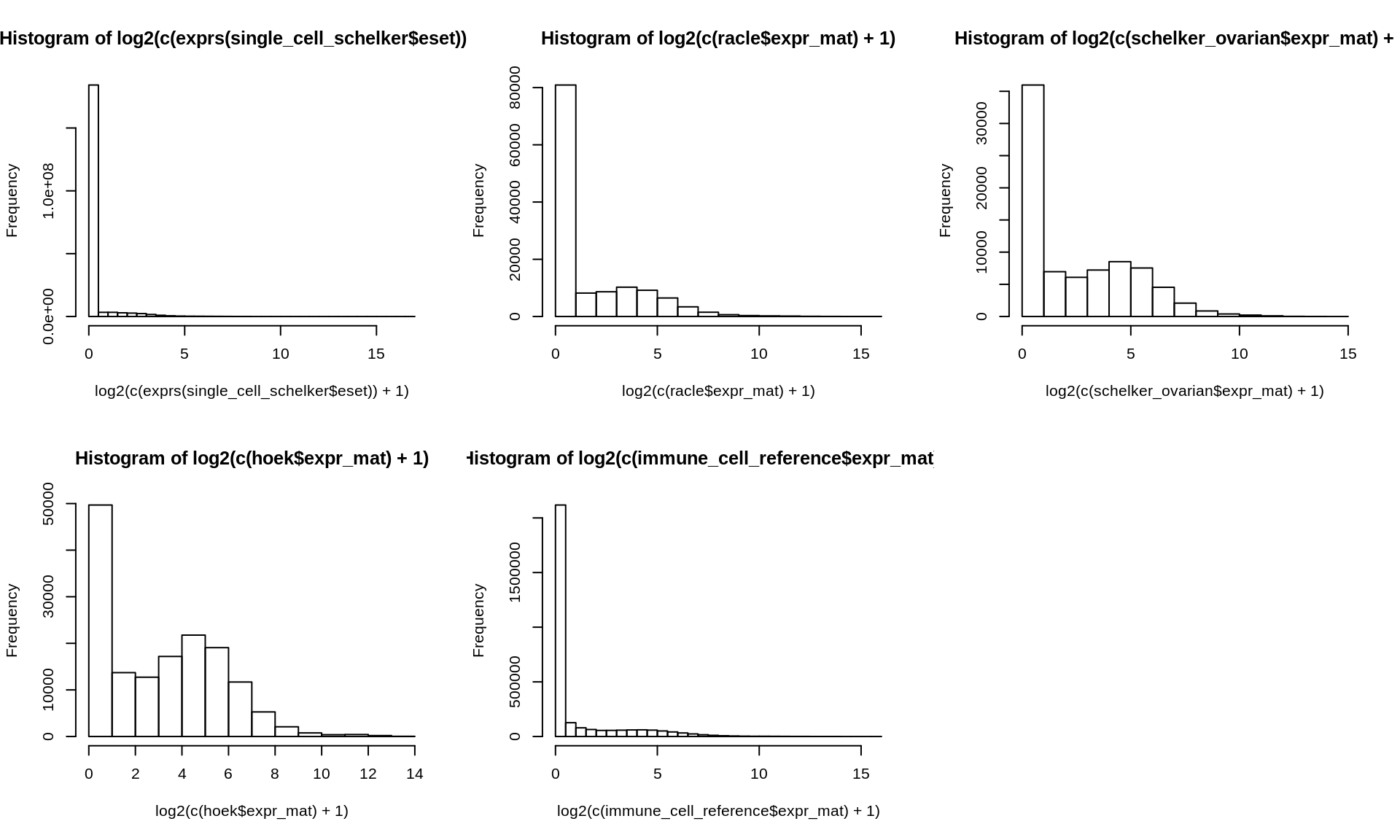
Figure 2.4: Histogram of log-tranformed gene expression data of all datasets
The mean values of all datasets:
mean(c(exprs(single_cell_schelker$eset)))
mean(c(racle$expr_mat))
mean(c(schelker_ovarian$expr_mat))
mean(c(hoek$expr_mat))
mean(c(immune_cell_reference$expr_mat))## [1] 0.7163828
## [1] 29.20031
## [1] 35.24549
## [1] 51.61823
## [1] 17.04449References
Schelker, Max, Sonia Feau, Jinyan Du, Nav Ranu, Edda Klipp, Gavin MacBeath, Birgit Schoeberl, and Andreas Raue. 2017. “Estimation of immune cell content in tumour tissue using single-cell RNA-seq data.” Nature Communications 8 (1): 2032. https://doi.org/10.1038/s41467-017-02289-3.
Finotello, Francesca, Clemens Mayer, Christina Plattner, Gerhard Laschober, Dietmar Rieder, Hubert Hackl, Anne Krogsdam, et al. 2017. “quanTIseq: quantifying immune contexture of human tumors.” bioRxiv, November. Cold Spring Harbor Laboratory, 223180. https://doi.org/10.1101/223180.
Hoek, Kristen L, Parimal Samir, Leigh M Howard, Xinnan Niu, Nripesh Prasad, Allison Galassie, Qi Liu, et al. 2015. “A Cell-Based Systems Biology Assessment of Human Blood to Monitor Immune Responses after Influenza Vaccination.” https://doi.org/10.1371/journal.pone.0118528.
Racle, Julien, Kaat de Jonge, Petra Baumgaertner, Daniel E Speiser, and David Gfeller. 2017. “Simultaneous enumeration of cancer and immune cell types from bulk tumor gene expression data.” eLife 6 (November). eLife Sciences Publications Limited: e26476. https://doi.org/10.7554/eLife.26476.